Research paper published in estimeed journal Nature Biotechnology
Allesøe, R.L., Lundgaard, A.T., Hernández Medina, R. et al. Discovery of drug–omics associations in type 2 diabetes with generative deep-learning models. Nature Biotechnol (2023). I.F., 68.164
Among the authors of the article is Prof. dr. Jerzy Adamski, since 2021 Visiting Scientist/Professor at the Faculty of Medicine of the University of Ljubljana in the Functional Genomics and Biotechnology for Health program group and in the Laboratory for the Study of Molecular Basis and Biomarkers of Hormone-Dependent Diseases of the Institute of Biochemistry and Molecular Genetics.
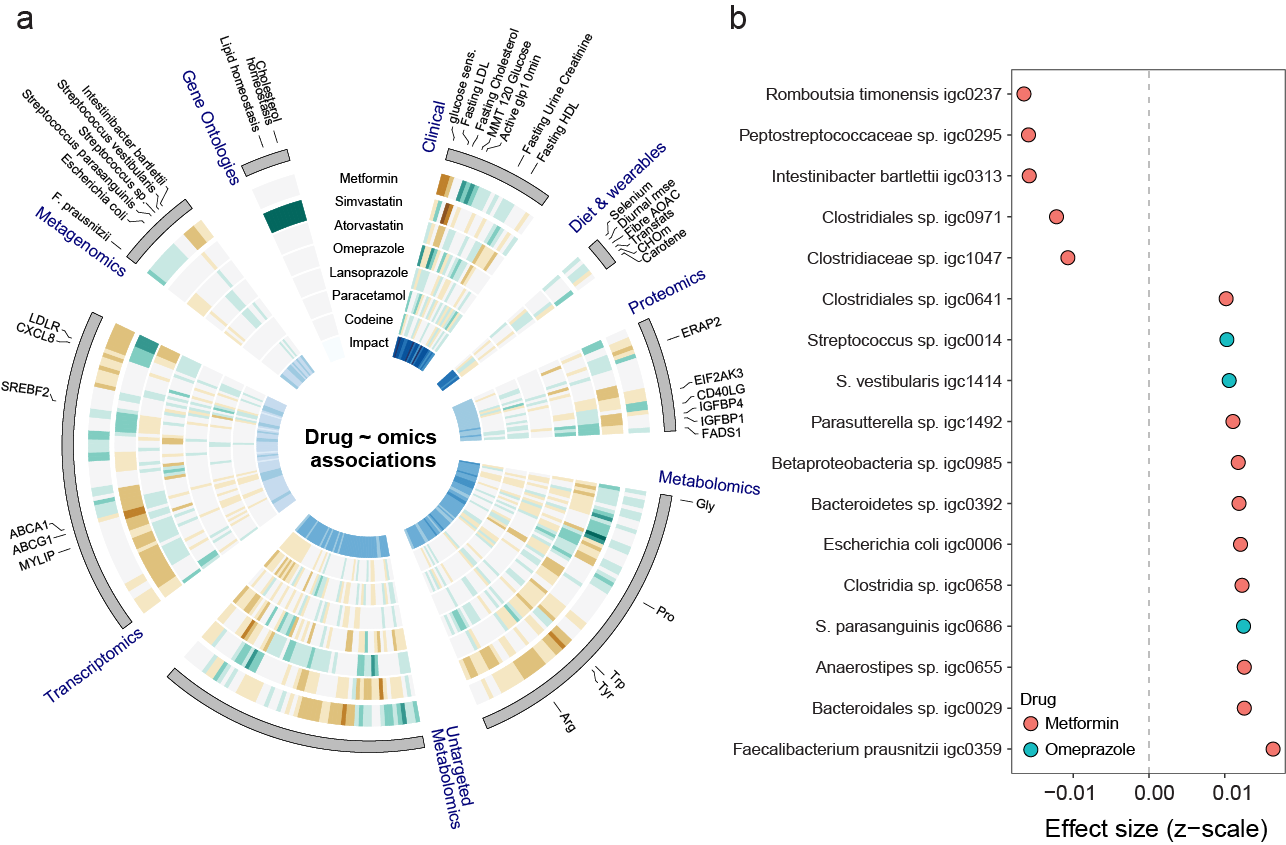
The application of multiple omics technologies in biomedical cohorts has the potential to reveal patient-level disease characteristics and individualized response to treatment. However, the scale and heterogeneous nature of multi-modal data makes integration and inference a non-trivial task. Group led by Prof. Rasmussen from the Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmarkdeveloped a deep-learning-based framework, and applied multi-omics variational autoencoders to integrate multi-modal data from a cohort of 789 people with newly diagnosed type 2 diabetes with deep multi-omics phenotyping done at the DIRECT consortium. They identifed drug–omics associations across the multi-modal datasets for the 20 most prevalent drugs given to people with type 2 diabetes. Among others, they identifed novel associations between metformin and the specific species of the gut microbiota as well as opposite molecular responses for the two statins, simvastatin and atorvastatin. They used the associations to quantify drug–drug similarities, assess the degree of polypharmacy. The described approach opens new possibilites in big multi-omics data analysis for discoveries of potential new biomarkers and investigating dirrect effects of drugs and more..